Homeobox
| Homeodomain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
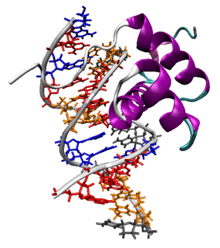 The Antennapedia homeodomain protein from Drosophila melanogaster bound to a fragment of DNA.[1] The recognition helix and unstructured N-terminus are bound in the major and minor grooves respectively. | |||||||||
| Identifiers | |||||||||
| Symbol | Homeodomain | ||||||||
| Pfam | PF00046 | ||||||||
| Pfam clan | CL0123 | ||||||||
| InterPro | IPR001356 | ||||||||
| SMART | SM00389 | ||||||||
| PROSITE | PDOC00027 | ||||||||
| SCOP | 1ahd | ||||||||
| SUPERFAMILY | 1ahd | ||||||||
| |||||||||
A homeobox is a DNA sequence, around 180 base pairs long, found within genes that are involved in the regulation of patterns of anatomical development (morphogenesis) in animals, fungi and plants. These genes encode homeodomain protein products that are transcription factors sharing a characteristic protein fold structure that binds DNA.[2][3][4] The "homeo-" prefix in the words "homeobox" and "homeodomain" stems from the mutational phenotype known as "homeosis", which is frequently observed when these genes are mutated in animals. Homeosis is a term coined by William Bateson to describe the outright replacement of a discrete body part with another body part.
Discovery
Homeoboxes were discovered independently in 1983 by Ernst Hafen, Michael Levine, and William McGinnis working in the lab of Walter Jakob Gehring at the University of Basel, Switzerland; and by Matthew P. Scott and Amy Weiner, who were then working with Thomas Kaufman at Indiana University in Bloomington.[5][6] The existence of homeobox genes were first discovered in Drosophila, where mutations in homeobox genes caused the radical alterations known as "homeotic transformations". One of the most famous such mutation is antennapedia, in which legs grow from the head of a fly instead of the expected antennae.
Homeodomain proteins
A homeobox is about 180 DNA base pairs long and encodes a protein domain that binds DNA. The following shows the consensus homeodomain (~60 amino acid residue chain), with typical insertion sites noted with dashes:[7]
RRRKRTA-YTRYQLLE-LEKEFLF-NRYLTRRRRIELAHSL-NLTERHIKIWFQNRRMKWKKEN
Structure
The characteristic homeodomain protein fold consists of a 60-amino acid helix-turn-helix (HTH) structure in which three alpha helices are connected by short loop regions. The N-terminal two helices are antiparallel and the longer C-terminal helix is roughly perpendicular to the axes established by the first two. It is this third helix that interacts directly with DNA via a number of hydrogen bonds and hydrophobic interactions, which occur between specific side chains and the exposed bases and thymine methyl groups within the major groove of the DNA.[8]
Homeodomain proteins are found in eukaryotes and prokaryotes[9] and have high homology to lambda phage proteins that alter the expression of genes in prokaryotes. The motif is very similar in sequence and structure in a wide range of DNA-binding proteins (e.g., cro and repressor proteins, homeotic proteins, etc.). One of the principal differences between HTH motifs in these different proteins arises from the stereo-chemical requirement for glycine in the turn which is needed to avoid steric interference of the beta-carbon with the main chain: for cro and repressor proteins the glycine appears to be mandatory, whereas for many of the homeotic and other DNA-binding proteins the requirement is relaxed.
Sequence specificity
Homeodomains can bind both specifically and nonspecifically to B-DNA with the C-terminal recognition helix aligning in the DNA's major groove and the unstructured peptide "tail" at the N-terminus aligning in the minor groove. The recognition helix and the inter-helix loops are rich in arginine and lysine residues, which form hydrogen bonds to the DNA backbone; conserved hydrophobic residues in the center of the recognition helix aid in stabilizing the helix packing. Homeodomain proteins show a preference for the DNA sequence 5'-ATTA-3'; sequence-independent binding occurs with significantly lower affinity.
Biological function
Through the DNA-recognition properties of the homeodomain, homeoproteins are believed to regulate the expression of targeted genes and direct the formation of many body structures during early embryonic development.[10] Many homeodomain proteins induce cellular differentiation by initiating the cascades of coregulated genes required to produce individual tissues and organs. Other proteins in the family, such as NANOG are involved in maintaining pluripotency. Homeobox genes are critical in the establishment of body axes during embryogenesis.
Homeoprotein transcription factors typically switch on cascades of other genes. The homeodomain binds DNA in a sequence-specific manner. However, the specificity of a single homeodomain protein is usually not enough to recognize only its desired target genes. Most of the time, homeodomain proteins act in the promoter region of their target genes as complexes with other transcription factors. Such complexes have a much higher target specificity than a single homeodomain protein. Homeodomains are encoded both by genes of the Hox gene clusters and by other genes throughout the genome.
The homeobox domain was first identified in a number of Drosophila homeotic and segmentation proteins, but is now known to be well-conserved in many other animals, including vertebrates.[2][8][11]
Specific members of the Hox family have been implicated in vascular remodeling, angiogenesis, and disease by orchestrating changes in matrix degradation, integrins, and components of the ECM.[12] HoxA5 is implicated in atherosclerosis.[13][14] HoxD3 and HoxB3 are proinvasive, angiogenic genes that upregulate b3 and a5 integrins and Efna1 in ECs, respectively.[15][16][17][18] HoxA3 induces endothelial cell (EC) migration by upregulating MMP14 and uPAR. Conversely, HoxD10 and HoxA5 have the opposite effect of suppressing EC migration and angiogenesis, and stabilizing adherens junctions by upregulating TIMP1/downregulating uPAR and MMP14, and by upregulating Tsp2/downregulating VEGFR2, Efna1, Hif1alpha and COX-2, respectively.[19][20] HoxA5 also upregulates the tumor suppressor p53 and Akt1 by downregulation of PTEN.[21] Suppression of HoxA5 has been shown to attenuate hemangioma growth.[22] HoxA5 has far-reaching effects on gene expression, causing ~300 genes to become upregulated upon its induction in breast cancer cell lines.[22] HoxA5 protein transduction domain overexpression prevents inflammation shown by inhibition of TNFalpha-inducible monocyte binding to HUVECs.[23][24][25]
Plant homeobox genes
As in animals, the plant homeobox genes code for the typical 60 amino acid long DNA-binding homeodomain or in case of the TALE (three amino acid loop extension) homeobox genes for an "atypical" homeodomain consisting of 63 amino acids. According to their conserved intron–exon structure and to unique codomain architectures they have been grouped into 14 distinct classes: HD-ZIP I to IV, BEL, KNOX, PLINC, WOX, PHD, DDT, NDX, LD, SAWADEE and PINTOX.[26] Conservation of codomains suggests a common eukaryotic ancestry for TALE[27] and non-TALE homeodomain proteins.[28]
POU genes
Proteins containing a POU region consist of a homeodomain and a separate, structurally homologous POU domain that contains two helix-turn-helix motifs and also binds DNA. The two domains are linked by a flexible loop that is long enough to stretch around the DNA helix, allowing the two domains to bind on opposite sides of the target DNA, collectively covering an eight-base segment with consensus sequence 5'-ATGCAAAT-3'. The individual domains of POU proteins bind DNA only weakly, but have strong sequence-specific affinity when linked. The POU domain itself has significant structural similarity with repressors expressed in bacteriophages, particularly lambda phage.
Human POU genes
HDX; POU1F1; POU2F1; POU2F2; POU2F3; POU3F1; POU3F2; POU3F3; POU3F4; POU4F1; POU4F2; POU4F3; POU5F1; POU5F1P1; POU5F1P4; POU5F2; POU6F1; POU6F2;
Hox genes
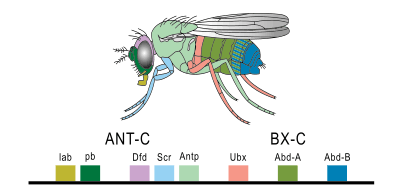
Hox genes are a subset of homeobox genes. They are essential metazoan genes as they determine the identity of embryonic regions along the anterio-posterior axis.[29] The first vertebrate Hox gene was isolated in Xenopus by Eddy De Robertis and colleagues in 1984, marking the beginning of the young science of evolutionary developmental biology ("evo-devo").[30]
In vertebrates, the four paralog clusters are partially redundant in function, but have also acquired several derived functions. In particular, HoxA and HoxD specify segment identity along the limb axis.
The main interest in this set of genes stems from their unique behaviour. They are typically found in an organized cluster. The linear order of the genes within a cluster is directly correlated to the order of the regions they affect as well as the timing in which they are affected. This phenomenon is called colinearity. Due to this linear relationship, changes in the gene cluster due to mutations generally result in similar changes in the affected regions.
For example, when one gene is lost the segment develops into a more anterior one, while a mutation that leads to a gain of function causes a segment to develop into a more posterior one. This is called ectopia. Famous examples are Antennapedia and bithorax in Drosophila, which can cause the development of legs instead of antennae and the development of a duplicated thorax, respectively.
Molecular evidence shows that some limited number of Hox genes have existed in the Cnidaria since before the earliest true Bilatera, making these genes pre-Paleozoic.[31] Homeobox genes have even been found in fungi, for example the unicellular yeasts, and in plants.
Human HOX genes
The Hox genes in humans are organized in four chromosomal clusters:
| name | chromosome | gene |
| HOXA (or sometimes HOX1) - HOXA@ | chromosome 7 | HOXA1, HOXA2, HOXA3, HOXA4, HOXA5, HOXA6, HOXA7, HOXA9, HOXA10, HOXA11, HOXA13 |
| HOXB - HOXB@ | chromosome 17 | HOXB1, HOXB2, HOXB3, HOXB4, HOXB5, HOXB6, HOXB7, HOXB8, HOXB9, HOXB13 |
| HOXC - HOXC@ | chromosome 12 | HOXC4, HOXC5, HOXC6, HOXC8, HOXC9, HOXC10, HOXC11, HOXC12, HOXC13 |
| HOXD - HOXD@ | chromosome 2 | HOXD1, HOXD3, HOXD4, HOXD8, HOXD9, HOXD10, HOXD11, HOXD12, HOXD13 |
Human DLX genes
There is also a "distal-less homeobox" family: DLX1, DLX2, DLX3, DLX4, DLX5, and DLX6. Dlx genes are involved in the development of the nervous system and of limbs.[32]
Human TALE genes
TALE (Three Amino acid Loop Extension) homeobox genes for an "atypical" homeodomain consisting of 63 rather than 60 amino acids. IRX1; IRX2; IRX3; IRX4; IRX5; IRX6; MEIS1; MEIS2; MEIS3; MKX; PBX1; PBX2; PBX3; PBX4; PKNOX1; PKNOX2;
Additional human homeobox genes and proteins containing this domain per UniProt annotation
- ADNP; ALX1 (CART1); ALX3; ALX4; ARGFX; ARX;
- BARHL1; BARHL2; BARX1; BARX2;
- CDX1; CDX2; CDX4; CRX; CUTL1; CUTL2;
- DBX1; DBX2; DMBX1; DPRX; DRGX; DUX1; DUX2; DUX3; DUX4; DUX4c; DUX5; DUXA;
- EMX1; EMX2; EN1; EN2; ESX1L; EVX1; EVX2;
- GBX1; GBX2; GSC; GSC2; GSX1; GSX2;
- HESX1; HHEX; HLX1; HMBOX1; HMX1; HMX2; HMX3; HNF1A; HNF1B; HOMEZ; HOPX
- ISL1; ISL2; ISX
- LASS2; LASS3; LASS4; LASS5; LASS6; LBX1; LBX2; LHX1; LHX2; LHX3; LHX4; LHX5; LHX6; LHX8; LHX9; LMX1A; LMX1B
- MEOX1; MEOX2; MIXL1; MNX1; MSX1; MSX2
- NANOG; NKX2-1; NKX2-2; NKX2-3; NKX2-4; NKX2-5; NKX2-8; NKX3-1; NKX3-2; NKX6-1; NKX6-2; NKX6-3; NOBOX; NOTO
- ONECUT1; ONECUT2; ONECUT3; OTP; OTX1; OTX2
- PAX3; PAX4; PAX6; PAX7; PDX1; PHOX2A; PHOX2B; PITX1; PITX2; PITX3; PROP1; PRRX1; PRRX2
- RAX; RAX2; RHOXF1; RHOXF2
- SATB1; SATB2; SEBOX; SHOX; SHOX2; SIX1; SIX2; SIX3; SIX4; SIX5; SIX6
- TGIF1; TGIF2; TGIF2LX; TGIF2LY; TLX1; TLX2; TLX3; TSHZ1; TSHZ2; TSHZ3
- UNCX; VAX1; VAX2; VENTX; VSX1; VSX2
- ZEB1; ZEB2; ZFHX2; ZFHX3; ZFHX4; ZHX1
Mutations
Mutations to homeobox genes can produce easily visible phenotypic changes.
Two examples of homeobox mutations in the above-mentioned fruit fly are legs where the antennae should be (antennapedia), and a second pair of wings.
Duplication of homeobox genes can produce new body segments, and such duplications are likely to have been important in the evolution of segmented animals. However, Hox genes typically determine the identity of body segments.
Interestingly, there is one insect family, the xyelid sawflies, in which both the antennae and mouthparts are remarkably leg-like in structure. This is not uncommon in arthropods as all arthropod appendages are homologous.
Regulation
Hox genes and their associated microRNAs are highly conserved developmental master regulators with tight tissue-specific, spatiotemporal control. These genes are known to be dysregulated in several cancers and are often controlled by DNA methylation.[13][33] The regulation of Hox genes is highly complex and involves reciprocal interactions, mostly inhibitory. Drosophila is known to use the Polycomb and Trithorax Complexes to maintain the expression of Hox genes after the down-regulation of the pair-rule and gap genes that occurs during larval development. Polycomb-group proteins can silence the HOX genes by modulation of chromatin structure.[34]
See also
References
- ↑ PDB: 1AHD; Billeter M, Qian YQ, Otting G, Müller M, Gehring W, Wüthrich K (Dec 1993). "Determination of the nuclear magnetic resonance solution structure of an Antennapedia homeodomain-DNA complex". Journal of Molecular Biology. 234 (4): 1084–93. doi:10.1006/jmbi.1993.1661. PMID 7903398.
- 1 2 Gehring WJ (Aug 1992). "The homeobox in perspective". Trends in Biochemical Sciences. 17 (8): 277–80. doi:10.1016/0968-0004(92)90434-B. PMID 1357790.
- ↑ Gehring WJ (Dec 1993). "Exploring the homeobox". Gene. 135 (1-2): 215–21. doi:10.1016/0378-1119(93)90068-E. PMID 7903947.
- ↑ Bürglin, TR, Affolter, M (Oct 2015). "Homeodomain proteins: an update". Chromosoma. in press (in press): 1–25. doi:10.1007/s00412-015-0543-8. PMID 26464018.
- ↑ McGinnis W, Levine MS, Hafen E, Kuroiwa A, Gehring WJ (1984). "A conserved DNA sequence in homoeotic genes of the Drosophila Antennapedia and bithorax complexes". Nature. 308 (5958): 428–33. doi:10.1038/308428a0. PMID 6323992.
- ↑ Scott MP, Weiner AJ (Jul 1984). "Structural relationships among genes that control development: sequence homology between the Antennapedia, Ultrabithorax, and fushi tarazu loci of Drosophila". Proceedings of the National Academy of Sciences of the United States of America. 81 (13): 4115–9. doi:10.1073/pnas.81.13.4115. PMC 345379
 . PMID 6330741.
. PMID 6330741. - ↑ Bürglin TR. "The homeobox page" (gif). Karolinksa Institute.
- 1 2 Schofield PN (1987). "Patterns, puzzles and paradigms - The riddle of the homeobox". Trends Neurosci. 10: 3–6. doi:10.1016/0166-2236(87)90113-5.
- ↑ http://www.cathdb.info/version/v4_0_0/superfamily/1.10.10.60
- ↑ Corsetti MT, Briata P, Sanseverino L, Daga A, Airoldi I, Simeone A, Palmisano G, Angelini C, Boncinelli E, Corte G (Sep 1992). "Differential DNA binding properties of three human homeodomain proteins". Nucleic Acids Research. 20 (17): 4465–72. doi:10.1093/nar/20.17.4465. PMC 334173
 . PMID 1357628.
. PMID 1357628. - ↑ Scott MP, Tamkun JW, Hartzell GW (Jul 1989). "The structure and function of the homeodomain". Biochimica et Biophysica Acta. 989 (1): 25–48. doi:10.1016/0304-419x(89)90033-4. PMID 2568852.
- ↑ Gorski DH, Walsh K (Nov 2000). "The role of homeobox genes in vascular remodeling and angiogenesis". Circulation Research. 87 (10): 865–72. doi:10.1161/01.res.87.10.865. PMID 11073881.
- 1 2 Dunn J, Thabet S, Jo H (Jul 2015). "Flow-Dependent Epigenetic DNA Methylation in Endothelial Gene Expression and Atherosclerosis". Arteriosclerosis, Thrombosis, and Vascular Biology. 35 (7): 1562–9. doi:10.1161/ATVBAHA.115.305042. PMID 25953647.
- ↑ Dunn J, Simmons R, Thabet S, Jo H (Oct 2015). "The role of epigenetics in the endothelial cell shear stress response and atherosclerosis". The International Journal of Biochemistry & Cell Biology. 67: 167–76. doi:10.1016/j.biocel.2015.05.001. PMID 25979369.
- ↑ Boudreau N, Andrews C, Srebrow A, Ravanpay A, Cheresh DA (Oct 1997). "Induction of the angiogenic phenotype by Hox D3". The Journal of Cell Biology. 139 (1): 257–64. doi:10.1083/jcb.139.1.257. PMID 9314544.
- ↑ Boudreau NJ, Varner JA (Feb 2004). "The homeobox transcription factor Hox D3 promotes integrin alpha5beta1 expression and function during angiogenesis". The Journal of Biological Chemistry. 279 (6): 4862–8. doi:10.1074/jbc.M305190200. PMID 14610084.
- ↑ Myers C, Charboneau A, Boudreau N (Jan 2000). "Homeobox B3 promotes capillary morphogenesis and angiogenesis". The Journal of Cell Biology. 148 (2): 343–51. doi:10.1083/jcb.148.2.343. PMC 2174277
 . PMID 10648567.
. PMID 10648567. - ↑ Chen Y, Xu B, Arderiu G, Hashimoto T, Young WL, Boudreau N, Yang GY (Nov 2004). "Retroviral delivery of homeobox D3 gene induces cerebral angiogenesis in mice". Journal of Cerebral Blood Flow and Metabolism. 24 (11): 1280–7. doi:10.1097/01.WCB.0000141770.09022.AB. PMID 15545924.
- ↑ Myers C, Charboneau A, Cheung I, Hanks D, Boudreau N (Dec 2002). "Sustained expression of homeobox D10 inhibits angiogenesis". The American Journal of Pathology. 161 (6): 2099–109. doi:10.1016/S0002-9440(10)64488-4. PMID 12466126.
- ↑ Mace KA, Hansen SL, Myers C, Young DM, Boudreau N (Jun 2005). "HOXA3 induces cell migration in endothelial and epithelial cells promoting angiogenesis and wound repair". Journal of Cell Science. 118 (Pt 12): 2567–77. doi:10.1242/jcs.02399. PMID 15914537.
- ↑ Rhoads K, Arderiu G, Charboneau A, Hansen SL, Hoffman W, Boudreau N (2005). "A role for Hox A5 in regulating angiogenesis and vascular patterning". Lymphatic Research and Biology. 3 (4): 240–52. doi:10.1089/lrb.2005.3.240. PMID 16379594.
- 1 2 Arderiu G, Cuevas I, Chen A, Carrio M, East L, Boudreau NJ. "HoxA5 stabilizes adherens junctions via increased Akt1". Cell Adhesion & Migration. 1 (4): 185–95. doi:10.4161/cam.1.4.5448. PMID 19262140.
- ↑ Zhu Y, Cuevas IC, Gabriel RA, Su H, Nishimura S, Gao P, Fields A, Hao Q, Young WL, Yang GY, Boudreau NJ (Jun 2009). "Restoring transcription factor HoxA5 expression inhibits the growth of experimental hemangiomas in the brain". Journal of Neuropathology and Experimental Neurology. 68 (6): 626–32. doi:10.1097/NEN.0b013e3181a491ce. PMID 19458547.
- ↑ Chen H, Rubin E, Zhang H, Chung S, Jie CC, Garrett E, Biswal S, Sukumar S (May 2005). "Identification of transcriptional targets of HOXA5". The Journal of Biological Chemistry. 280 (19): 19373–80. doi:10.1074/jbc.M413528200. PMID 15757903.
- ↑ Lee JY, Park KS, Cho EJ, Joo HK, Lee SK, Lee SD, Park JB, Chang SJ, Jeon BH (Jul 2011). "Human HOXA5 homeodomain enhances protein transduction and its application to vascular inflammation". Biochemical and Biophysical Research Communications. 410 (2): 312–6. doi:10.1016/j.bbrc.2011.05.139. PMID 21664342.
- ↑ Mukherjee K, Brocchieri L, Bürglin TR (Dec 2009). "A comprehensive classification and evolutionary analysis of plant homeobox genes". Molecular Biology and Evolution. 26 (12): 2775–94. doi:10.1093/molbev/msp201. PMC 2775110
 . PMID 19734295.
. PMID 19734295. - ↑ Bürglin TR (Nov 1997). "Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals". Nucleic Acids Research. 25 (21): 4173–80. doi:10.1093/nar/25.21.4173. PMC 147054
 . PMID 9336443.
. PMID 9336443. - ↑ Derelle R, Lopez P, Le Guyader H, Manuel M (2007). "Homeodomain proteins belong to the ancestral molecular toolkit of eukaryotes". Evolution & Development. 9 (3): 212–9. doi:10.1111/j.1525-142X.2007.00153.x. PMID 17501745.
- ↑ Alonso CR (Nov 2002). "Hox proteins: sculpting body parts by activating localized cell death". Current Biology. 12 (22): R776–8. doi:10.1016/S0960-9822(02)01291-5. PMID 12445403.
- ↑ Carrasco AE, McGinnis W, Gehring WJ, De Robertis EM (Jun 1984). "Cloning of an X. laevis gene expressed during early embryogenesis coding for a peptide region homologous to Drosophila homeotic genes". Cell. 37 (2): 409–14. doi:10.1016/0092-8674(84)90371-4. PMID 6327066.
- ↑ Ryan JF, Mazza ME, Pang K, Matus DQ, Baxevanis AD, Martindale MQ, Finnerty JR (2007). "Pre-bilaterian origins of the Hox cluster and the Hox code: evidence from the sea anemone, Nematostella vectensis". PLOS ONE. 2 (1): e153. doi:10.1371/journal.pone.0000153. PMC 1779807
 . PMID 17252055.
. PMID 17252055. - ↑ Kraus P, Lufkin T (Jul 2006). "Dlx homeobox gene control of mammalian limb and craniofacial development". American Journal of Medical Genetics Part A. 140 (13): 1366–74. doi:10.1002/ajmg.a.31252. PMID 16688724.
- ↑ Bhatlekar S, Fields JZ, Boman BM (Aug 2014). "HOX genes and their role in the development of human cancers". Journal of Molecular Medicine. 92 (8): 811–23. doi:10.1007/s00109-014-1181-y. PMID 24996520.
- ↑ Portoso M, Cavalli G (2008). "The Role of RNAi and Noncoding RNAs in Polycomb Mediated Control of Gene Expression and Genomic Programming". RNA and the Regulation of Gene Expression: A Hidden Layer of Complexity. Caister Academic Press. ISBN 978-1-904455-25-7.
Further reading
- Lodish H, Berk A, Matsudaira P, Kaiser CA, Krieger M, Scott MP, Zipursky L, Darnell J (2003). Molecular Cell Biology (5th ed.). New York: W.H. Freeman and Company. ISBN 978-0-7167-4366-8.
- Tooze C, Branden J (1999). Introduction to protein structure (2nd ed.). New York: Garland Pub. pp. 159–66. ISBN 978-0-8153-2305-1.
- Ogishima S, Tanaka H (Jan 2007). "Missing link in the evolution of Hox clusters". Gene. 387 (1-2): 21–30. doi:10.1016/j.gene.2006.08.011. PMID 17098381.
External links
- The Homeodomain Resource (National Human Genome Research Institute, National Institutes of Health)
- HomeoDB: a database of homeobox genes diversity. Zhong YF, Butts T, Holland PWH, since 2008.
- Eukaryotic Linear Motif resource motif class LIG_HOMEOBOX
- Homeobox at the US National Library of Medicine Medical Subject Headings (MeSH)
This article incorporates text from the public domain Pfam and InterPro IPR001356