ETS1
| View/Edit Human | View/Edit Mouse |
Protein C-ets-1 is a protein that in humans is encoded by the ETS1 gene.[4] The protein encoded by this gene belongs to the ETS family of transcription factors.[5]
Function
There are 28 ETS genes in humans and 27 genes – in mice. They bind the DNA via their winged-helix-turn-helix DNA binding motif known as the Ets domain that specifically recognizes DNA sequences that contain a GGAA/T core element. However, Ets proteins differ significantly in their preference for the sequence flanking the GGAA/T core motif. For instance, the consensus sequence for Ets1 is PuCC/a-GGAA/T-GCPy. On the other hand, many natural Ets1-responsive GGAA/T elements differ from this consensus sequence. The later suggests that several other transcription factors may facilitate Ets1 binding to unfavorable DNA sequences.
Ets1 binds to DNA as a monomer. Phosphorylation of serine residues of the C-terminal domain (in the nucleotide sequence they belong to exon VII) known as autoinhibition makes Ets1 inactive. There are several ways to activate Ets1. First, Ets1 can be dephosphorylated. Second, two Ets1 can be activated If two Ets molecules homodimerize. The homodimerization occurs if DNA binding sites are present in the correct orientation and spacing. Thus, the exact layout of binding sites within an enhancer or promoter segment to either relieve or allow autoinhibition of Ets1 to occur may strongly influence whether or not Ets1 actually binds to particular site. Third, Ets1 can be activated by Erk2 and Ras at Thr38. The truncated isoform cannot be phosphorylated by the Erk2. It is localized in the cytoplasm and acts as a dominant negative isoform. Contrary, another isoform that misses exon VII is constitutively active. Many Ras responsive genes harbor combinatorial Ets/AP1 recognition motifs through which Ets1 and AP1 synergistically activate transcription when stimulated by Ras.[6]
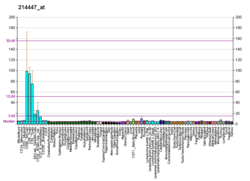
In adult humans, Ets1 is expressed at high levels mainly in immune tissues such as thymus, spleen, and lymph node (B cells, T cells, NK cells, and NK T cells and non-lymphoid immune cells). An enforced expression of Ets1 blocks differentiation of B- and T-cells. Contrary, knocking Ets1 down is causing multiple defects in the immune system.
Knockout mice
Ets1 knockout mice have aberrant thymic differentiation, reduced peripheral T cell numbers, reduced IL-2 production, a skewing towards a memory/effector phenotype and impairments in the production of Th1 and Th2 cytokines. Although Ets1 knockout mice have an impaired development of Th1, Th2, and Treg cells, they have higher numbers of Th17 cells. There are also partial defects in bone marrow B cell development with reduced cellularity and inefficient transition from pro-B to pre-B cell stages.
Clinical significance
Meta-analyses of multiple genome-wide association studies has suggested an association of SNPs in the ETS1locus with psoriasis in European populations. This is not surprising because Ets1 is a negative regulator of Th17 cells.
Ets1 overexpression in stratified squamous epithelial cells causes pro-oncogenic changes, such as suspension of terminal differentiation, high secretion of matrix metalloproteases (Mmps), epidermal growth factor ligands, and inflammatory mediators.
Interactions
Ets1 directly interacts with various transcription factors. Their interaction results in formation of multiprotein complexes. When Ets1 interacts with other transcription factors (Runx1, Pax5, TFE3, and USF1) its final effect on transcription depends on whether C-terminal domain is phosphorylated. Acetyltransferases CBP and p300 bind to the transactivation domain. AP1, STAT5 and VDR bind to C-terminal domain.
Also, ETS1 has been shown to interact with TTRAP,[7] UBE2I[8] and Death associated protein 6.[9]
References
- ↑ "Diseases that are genetically associated with ETS1 view/edit references on wikidata".
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, Kovar H, Joubert I, de Jong P, Rouleau G (Sep 1992). "Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours". Nature. 359 (6391): 162–5. doi:10.1038/359162a0. PMID 1522903.
- ↑ Dwyer J, Li H, Xu D, Liu JP (Oct 2007). "Transcriptional regulation of telomerase activity: roles of the Ets transcription factor family". Annals of the New York Academy of Sciences. 1114 (1): 36–47. doi:10.1196/annals.1396.022. PMID 17986575.
- ↑ Wasylyk B, Hagman J, Gutierrez-Hartmann A (1998). "Ets transcription factors: nuclear effectors of the Ras-MAP-kinase signaling pathway". Trends Biochem. Sci. 23 (6): 213–6. doi:10.1016/S0968-0004(98)01211-0. PMID 9644975.
- ↑ Pei H, Yordy JS, Leng Q, Zhao Q, Watson DK, Li R (May 2003). "EAPII interacts with ETS1 and modulates its transcriptional function". Oncogene. 22 (18): 2699–709. doi:10.1038/sj.onc.1206374. PMID 12743594.
- ↑ Hahn SL, Wasylyk B, Criqui-Filipe P, Criqui P (Sep 1997). "Modulation of ETS-1 transcriptional activity by huUBC9, a ubiquitin-conjugating enzyme". Oncogene. 15 (12): 1489–95. doi:10.1038/sj.onc.1201301. PMID 9333025.
- ↑ Li R, Pei H, Watson DK, Papas TS (Feb 2000). "EAP1/Daxx interacts with ETS1 and represses transcriptional activation of ETS1 target genes". Oncogene. 19 (6): 745–53. doi:10.1038/sj.onc.1203385. PMID 10698492.
Further reading
- Reddy ES, Rao VN (1988). "Structure, expression and alternative splicing of the human c-ets-1 proto-oncogene". Oncogene Research. 3 (3): 239–46. PMID 3060801.
- Lincoln DW, Bove K (Jan 2005). "The transcription factor Ets-1 in breast cancer". Frontiers in Bioscience. 10 (1-3): 506–11. doi:10.2741/1546. PMID 15574387.
External links
- ETS1 protein, human at the US National Library of Medicine Medical Subject Headings (MeSH)
- Drosophila pointed - The Interactive Fly
- FactorBook ETS1
This article incorporates text from the United States National Library of Medicine, which is in the public domain.