A-DNA
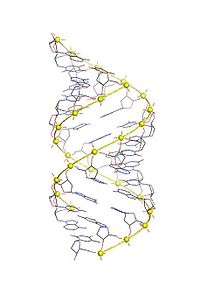
A-DNA is one of the possible double helical structures which DNA can adopt. A-DNA is thought to be one of three biologically active double helical structures along with B-DNA and Z-DNA. It is a right-handed double helix fairly similar to the more common B-DNA form, but with a shorter, more compact helical structure whose base pairs are not perpendicular to the helix-axis as in B-DNA. It was discovered by Rosalind Franklin, who also named the A and B forms. She showed that DNA is driven into the A form when under dehydrating conditions. Such conditions are commonly used in to form crystals, and many DNA crystal structures are in the A form. The same helical conformation occurs in double-stranded RNAs, and in DNA-RNA hybrid double helices.
Structure
A-DNA is fairly similar to B-DNA given that it is a right-handed double helix with major and minor grooves. However, as shown in the comparison table below, there is a slight increase in the number of base pairs (bp) per turn (resulting in a smaller twist angle), and smaller rise per base pair (making A-DNA 20-25% shorter than B-DNA). The major groove of A-DNA is deep and narrow, while the minor groove is wide and shallow.
Comparison geometries of the most common DNA forms
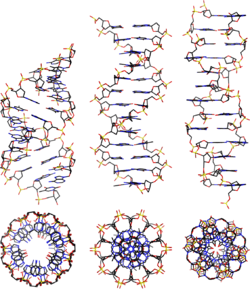
| Geometry attribute: | A-form | B-form | Z-form |
|---|---|---|---|
| Helix sense | right-handed | right-handed | left-handed |
| Repeating unit | 1 bp | 1 bp | 2 bp |
| Rotation/bp | 32.7° | 34.3° | 60°/2 |
| Mean bp/turn | 11 | 10.5 | 12 |
| Inclination of bp to axis | +19° | −1.2° | −9° |
| Rise/bp along axis | 2.6 Å (0.26 nm) | 3.4 Å (0.34 nm) | 3.7 Å (0.37 nm) |
| Rise/turn of helix | 28.6 Å (2.86 nm) | 35.7 Å (3.57 nm) | 45.6 Å (4.56 nm) |
| Mean propeller twist | +18° | +16° | 0° |
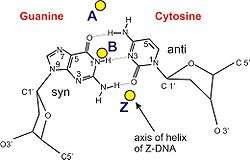
| Glycosyl angle | anti | anti | pyrimidine: anti, purine: syn |
| Nucleotide phosphate to phosphate distance | 5.9 Å | 7.0 Å | C: 5.7 Å, G: 6.1 Å |
| Sugar pucker | C3'-endo | C2'-endo | C: C2'-endo, G: C3'-endo |
| Diameter | 23 Å (2.3 nm) | 20 Å (2.0 nm) | 18 Å (1.8 nm) |
Biological function
Dehydration of DNA drives it into the A form, and this apparently protects DNA under conditions such as the extreme desiccation of bacteria.[1] Protein binding can also strip solvent off of DNA and convert it to the A form, as revealed by the structure of a rod-shaped virus.[2]
It has been proposed that the motors that package double-stranded DNA in bacteriophages exploit the fact that A-DNA is shorter than B-DNA, and that conformational changes in the DNA itself are the source of the large forces generated by these motors.[3] In this model, ATP hydrolysis is used to drive protein conformational changes that alternatively dehydrate and rehydrate the DNA, and the DNA shortening/lengthening cycle is coupled to a protein-DNA grip/release cycle to generate the forward motion that moves DNA into the capsid.
See also
References
- ↑ Whelan DR, et al. (2014). "Detection of an en masse and reversible B- to A-DNA conformational transition in prokaryotes in response to desiccation". J R Soc Interface. 11: 20140454. doi:10.1098/rsif.2014.0454. PMID 24898023.
- ↑ Di Maio F, Egelman EH, et al. (2015). "A virus that infects a hyperthermophile encapsidates A-form DNA". Science. 348: 914–917. doi:10.1126/science.aaa4181. PMID 25999507.
- ↑ Harvey, SC (2015). "The scrunchworm hypothesis: Transitions between A-DNA and B-DNA provide the driving force for genome packaging in double-stranded DNA bacteriophages". Journal of Structural Biology. 189: 1–8. doi:10.1016/j.jsb.2014.11.012. PMID 25486612.