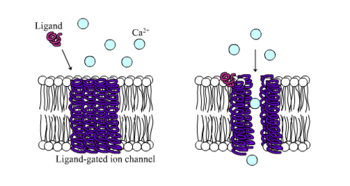
Ligand-gated ion channel
| Neurotransmitter-gated ion-channel transmembrane region | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Ligand-gated ion channel | |||||||||
| Identifiers | |||||||||
| Symbol | Neur_chan_memb | ||||||||
| Pfam | PF02932 | ||||||||
| InterPro | IPR006029 | ||||||||
| PROSITE | PDOC00209 | ||||||||
| SCOP | 1cek | ||||||||
| SUPERFAMILY | 1cek | ||||||||
| TCDB | 1.A.9 | ||||||||
| OPM superfamily | 14 | ||||||||
| OPM protein | 2bg9 | ||||||||
| |||||||||
Ligand-gated ion channels (LICs), (TC# 1.A.9), also commonly referred as ionotropic receptors, are a group of transmembrane ion channel proteins which open to allow ions such as Na+, K+, Ca2+, and/or Cl− to pass through the membrane in response to the binding of a chemical messenger (i.e. a ligand),[1] such as a neurotransmitter.[2]
These proteins are typically composed of at least two different domains: a transmembrane domain which includes the ion pore, and an extracellular domain which includes the ligand binding location (an allosteric binding site). This modularity has enabled a 'divide and conquer' approach to finding the structure of the proteins (crystallising each domain separately). The function of such receptors located at synapses is to convert the chemical signal of presynaptically released neurotransmitter directly and very quickly into a postsynaptic electrical signal. Many LICs are additionally modulated by allosteric ligands, by channel blockers, ions, or the membrane potential. LICs are classified into three superfamilies which lack evolutionary relationship: cys-loop receptors, ionotropic glutamate receptors and ATP-gated channels.
Cys-loop receptors
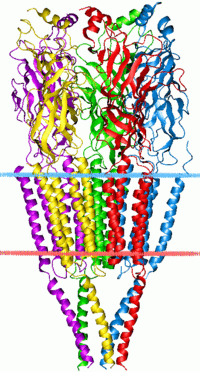
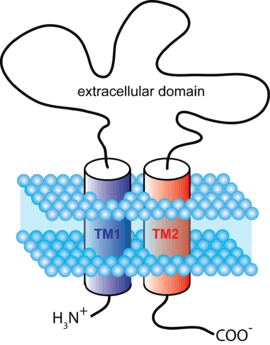
The cys-loop receptors are named after a characteristic loop formed by a disulfide bond between two cysteine residues in the N terminal extracellular domain. Because of this extracellular N-terminal ligand-binding domain, they exhibit receptor specificity for (1) acetylcholine (AcCh), (2) serotonin, (3) glycine, (4) glutamate and (5) γ-aminobutyric acid (GABA) in vertebrates. The receptors are subdivided with respect to the type of ion that they conduct (anionic or cationic) and further into families defined by the endogenous ligand. They are usually pentameric with each subunit containing 4 transmembrane helices constituting the transmembrane domain, and a beta sheet sandwich type, extracellular, N terminal, ligand binding domain.[3] Some also contain an intracellular domain like shown in the image.
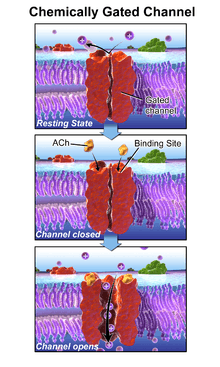
The prototypic ligand-gated ion channel is the nicotinic acetylcholine receptor. It consists of a pentamer of protein subunits (typically ααβγδ), with two binding sites for acetylcholine (one at the interface of each alpha subunit). When the acetylcholine binds it alters the receptor's configuration (twists the T2 helices which moves the leucine residues, which block the pore, out of the channel pathway) and causes the constriction in the pore of approximately 3 angstroms to widen to approximately 8 angstroms so that ions can pass through. This pore allows Na+ ions to flow down their electrochemical gradient into the cell. With a sufficient number of channels opening at once, the inward flow of positive charges carried by Na+ ions depolarizes the postsynaptic membrane sufficiently to initiate an action potential.
While single-cell organisms like bacteria would have little apparent need for the transmission of an action potential, a bacterial homologue to an LIC has been identified, hypothesized to act nonetheless as a chemoreceptor.[4] This prokaryotic nAChR variant is known as the GLIC receptor, after the species in which it was identified; Gloeobacter Ligand-gated Ion Channel.
Structure
Cys-loop receptors have structural elements that are well conserved, with a large extracellular domain (ECD) harboring an alpha-helix and 10 beta-strands. Following the ECD, four transmembrane segments (TMSs) are connected by intracellular and extracellular loop structures.[5] Except the TMS 3-4 loop, their lengths are only 7-14 residues. The TMS 3-4 loop forms the largest part of the intracellular domain (ICD) and exhibits the most variable region between all of these homologous receptors. The ICD is defined by the TMS 3-4 loop together with the TMS 1-2 loop preceding the ion channel pore.[5] Crystallization has revealed structures for some members of the family, but to allow crystallization, the intracellular loop was usually replaced by a short linker present in prokaryotic cys-loop receptors, so their structures as not known. Nevertheless, this intracellular loop appears to function in desensitization, modulation of channel physiology by pharmacological substances, and posttranslational modifications. Motifs important for trafficking are therein, and the ICD interacts with scaffold proteins enabling inhibitory synapse formation.[5]
Transport Reaction
The reaction catalyzed by LIC family members is:
- ions (in) ↔ ions (out)
Vertebrate Anionic Cys-loop Receptors
| Type | Class | IUPHAR-recommended protein name[6] |
Gene | Previous names |
|---|---|---|---|---|
| GABAA | alpha | α1 α2 α3 α4 α5 α6 |
GABRA1 GABRA2 GABRA3 GABRA4 GABRA5 GABRA6 |
EJM, ECA4 |
| beta | β1 β2 β3 |
GABRB1 GABRB2 GABRB3 |
ECA5 | |
| gamma | γ1 γ2 γ3 |
GABRG1 GABRG2 GABRG3 |
CAE2, ECA2, GEFSP3 | |
| delta | δ | GABRD | ||
| epsilon | ε | GABRE | ||
| pi | π | GABRP | ||
| theta | θ | GABRQ | ||
| rho | ρ1 ρ2 ρ3 |
GABRR1 GABRR2 GABRR3 |
GABAC[7] | |
| Glycine (GlyR) |
alpha | α1 α2 α3 α4 |
GLRA1 GLRA2 GLRA3 GLRA4 |
STHE |
| beta | β | GLRB |
Vertebrate Cationic Cys-loop Receptors
| Type | Class | IUPHAR-recommended protein name [6] |
Gene | Previous names |
|---|---|---|---|---|
| Serotonin (5-HT) |
5-HT3 | 5-HT3A 5-HT3B 5-HT3C 5-HT3D 5-HT3E |
HTR3A HTR3B HTR3C HTR3D HTR3E |
5-HT3A 5-HT3B 5-HT3C 5-HT3D 5-HT3E |
| Nicotinic acetylcholine (nAChR) |
alpha | α1 α2 α3 α4 α5 α6 α7 α9 α10 |
CHRNA1 CHRNA2 CHRNA3 CHRNA4 CHRNA5 CHRNA6 CHRNA7 CHRNA9 CHRNA10 |
ACHRA, ACHRD, CHRNA, CMS2A, FCCMS, SCCMS |
| beta | β1 β2 β3 β4 |
CHRNB1 CHRNB2 CHRNB3 CHRNB4 |
CMS2A, SCCMS, ACHRB, CHRNB, CMS1D EFNL3, nAChRB2 | |
| gamma | γ | CHRNG | ACHRG | |
| delta | δ | CHRND | ACHRD, CMS2A, FCCMS, SCCMS | |
| epsilon | ε | CHRNE | ACHRE, CMS1D, CMS1E, CMS2A, FCCMS, SCCMS | |
| Zinc-activated ion channel (ZAC) |
ZAC | ZACN | ZAC1, L2m LICZ, LICZ1 |
Ionotropic glutamate receptors (iGluR)
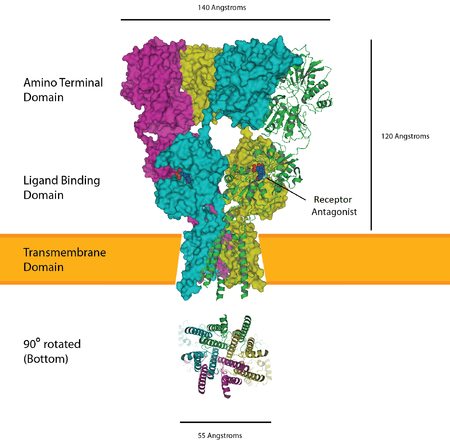
The ionotropic glutamate receptors bind the neurotransmitter glutamate. They form tetramers with each subunit consisting of an extracellular amino terminal domain (ATD, which is involved tetramer assembly), an extracellular ligand binding domain (LBD, which binds glutamate), and a transmembrane domain (TMD, which forms the ion channel). The transmembrane domain of each subunit contains three transmembrane helices as well as a half membrane helix with a reentrant loop. The structure of the protein starts with the ATD at the N terminus followed by the first half of the LBD which is interrupted by helices 1,2 and 3 of the TMD before continuing with the final half of the LBD and then finishing with helix 4 of the TMD at the C terminus. This means there are three links between the TMD and the extracellular domains. Each subunit of the tetramer has a binding site for glutamate formed by the two LBD sections forming a clamshell like shape. Only two of these sites in the tetramer need to be occupied to open the ion channel. The pore is mainly formed by the half helix 2 in a way which resembles an inverted potassium channel.
| Type | Class | IUPHAR-recommended protein name [6] |
Gene | Previous names |
|---|---|---|---|---|
| AMPA | GluA | GluA1 GluA2 GluA3 GluA4 |
GRIA1 GRIA2 GRIA3 GRIA4 |
GLUA1, GluR1, GluRA, GluR-A, GluR-K1, HBGR1 GLUA2, GluR2, GluRB, GluR-B, GluR-K2, HBGR2 GLUA3, GluR3, GluRC, GluR-C, GluR-K3 GLUA4, GluR4, GluRD, GluR-D |
| Kainate | GluK | GluK1 GluK2 GluK3 GluK4 GluK5 |
GRIK1 GRIK2 GRIK3 GRIK4 GRIK5 |
GLUK5, GluR5, GluR-5, EAA3 GLUK6, GluR6, GluR-6, EAA4 GLUK7, GluR7, GluR-7, EAA5 GLUK1, KA1, KA-1, EAA1 GLUK2, KA2, KA-2, EAA2 |
| NMDA | GluN | GluN1 NRL1A NRL1B |
GRIN1 GRINL1A GRINL1B |
GLUN1, NMDA-R1, NR1, GluRξ1 |
| GluN2A GluN2B GluN2C GluN2D |
GRIN2A GRIN2B GRIN2C GRIN2D |
GLUN2A, NMDA-R2A, NR2A, GluRε1 GLUN2B, NMDA-R2B, NR2B, hNR3, GluRε2 GLUN2C, NMDA-R2C, NR2C, GluRε3 GLUN2D, NMDA-R2D, NR2D, GluRε4 | ||
| GluN3A GluN3B |
GRIN3A GRIN3B |
GLUN3A, NMDA-R3A, NMDAR-L, chi-1 GLU3B, NMDA-R3B | ||
| ‘Orphan’ | (GluD) | GluD1 GluD2 |
GRID1 GRID2 |
GluRδ1 GluRδ2 |
ATP-gated channels
ATP-gated channels open in response to binding the nucleotide ATP. They form trimers with two transmembrane helices per subunit and both the C and N termini on the intracellular side.
| Type | Class | IUPHAR-recommended protein name [6] |
Gene | Previous names |
|---|---|---|---|---|
| P2X | N/A | P2X1 P2X2 P2X3 P2X4 P2X5 P2X6 P2X7 |
P2RX1 P2RX2 P2RX3 P2RX4 P2RX5 P2RX6 P2RX7 |
P2X1 P2X2 P2X3 P2X4 P2X5 P2X6 P2X7 |
PIP2-gated channels
Phosphatidylinositol 4,5-bisphosphate (PIP2) binds to and directly agonizes Inward rectifying potassium channels(Kir).[8] PIP2 is a plasma membrane lipid and its definitive role in gating ion channels was only recently demonstrated by X-ray crystallography.
Clinical relevance
Ligand-gated ion channels are likely to be the major site at which anaesthetic agents and ethanol have their effects, although unequivocal evidence of this is yet to be established.[9][10] In particular, the GABA and NMDA receptors are affected by anaesthetic agents at concentrations similar to those used in clinical anaesthesia.[11]
See also
References
- ↑ "ligand-gated channel" at Dorland's Medical Dictionary
- ↑ Purves, Dale, George J. Augustine, David Fitzpatrick, William C. Hall, Anthony-Samuel LaMantia, James O. McNamara, and Leonard E. White (2008). Neuroscience. 4th ed. Sinauer Associates. pp. 156–7. ISBN 978-0-87893-697-7.
- ↑ Cascio M (2004). "Structure and function of the glycine receptor and related nicotinicoid receptors". J. Biol. Chem. 279 (19): 19383–6. doi:10.1074/jbc.R300035200. PMID 15023997.
- ↑ Tasneem A, Iyer L, Jakobsson E, Aravind L (2004). "Identification of the prokaryotic ligand-gated ion channels and their implications for the mechanisms and origins of animal Cys-loop ion channels". Genome Biology. 6 (1): R4. doi:10.1186/gb-2004-6-1-r4. PMC 549065
 . PMID 15642096.
. PMID 15642096. - 1 2 3 Langlhofer, Georg; Villmann, Carmen (2016-01-01). "The Intracellular Loop of the Glycine Receptor: It's not all about the Size". Frontiers in Molecular Neuroscience. 9: 41. doi:10.3389/fnmol.2016.00041. ISSN 1662-5099. PMC 4891346
 . PMID 27330534.
. PMID 27330534. - 1 2 3 4 Collingridge GL, Olsen RW, Peters J, Spedding M (January 2009). "A nomenclature for ligand-gated ion channels". Neuropharmacology. 56 (1): 2–5. doi:10.1016/j.neuropharm.2008.06.063. PMC 2847504
 . PMID 18655795.
. PMID 18655795. - ↑ Olsen RW, Sieghart W (September 2008). "International Union of Pharmacology. LXX. Subtypes of γ-Aminobutyric AcidA Receptors: Classification on the Basis of Subunit Composition, Pharmacology, and Function. Update". Pharmacol. Rev. 60 (3): 243–60. doi:10.1124/pr.108.00505. PMC 2847512
 . PMID 18790874.
. PMID 18790874. - ↑ Hansen SB, Tao X, MacKinnon R (September 2011). "Structural basis of PIP2 activation of the classical inward rectifier K+ channel Kir2.2". Nature. 477 (7365): 495–8. Bibcode:2011Natur.477..495H. doi:10.1038/nature10370. PMC 3324908
 . PMID 21874019.
. PMID 21874019. - ↑ Krasowski MD, Harrison NL (1999). "General anaesthetic actions on ligand-gated ion channels". Cell. Mol. Life Sci. 55 (10): 1278–303. doi:10.1007/s000180050371. PMC 2854026
 . PMID 10487207.
. PMID 10487207. - ↑ Dilger JP (2002). "The effects of general anaesthetics on ligand-gated ion channels". Br J Anaesth. 89 (1): 41–51. doi:10.1093/bja/aef161. PMID 12173240.
- ↑ Harris RA, Mihic SJ, Dildy-Mayfield JE, Machu TK (1995). "Actions of anesthetics on ligand-gated ion channels: role of receptor subunit composition" (abstract). FASEB J. 9 (14): 1454–62. PMID 7589987.
External links
- Ligand-Gated Ion Channel database at European Bioinformatics Institute. Verified availability April 11, 2007.
- "Revised Recommendations for Nomenclature of Ligand-Gated Ion Channels". IUPHAR Database of Receptors and Ion Channels. International Union of Basic and Clinical Pharmacology.
As of this edit, this article uses content from "1.A.9 The Neurotransmitter Receptor, Cys loop, Ligand-gated Ion Channel (LIC) Family", which is licensed in a way that permits reuse under the Creative Commons Attribution-ShareAlike 3.0 Unported License, but not under the GFDL. All relevant terms must be followed.