Isogenic human disease models
Isogenic human disease models are a family of cells that are selected or engineered to accurately model the genetics of a specific patient population, in vitro. They are provided with a genetically matched ‘normal cell’ to provide an isogenic system to research disease biology and novel therapeutic agents.[1] They can be used to model any disease with a genetic foundation. Cancer is one such disease for which isogenic human disease models have been widely used.
Historical models
Human isogenic disease models have been likened to ‘patients in a test-tube’, since they incorporate the latest research into human genetic diseases and do so without the difficulties and limitations involved in using non-human models.[2]
Historically, cells obtained from animals, typically mice, have been used to model cancer related pathways. However, there are obvious limitations inherent in using animals for modelling genetically determined diseases in humans. Despite a large proportion of genetic conservation between humans and mice, there are significant differences between the biology of mice and humans that are important to cancer research. For example, major differences in telomere regulation enable murine cells to bypass the requirement for telomerase upregulation, which is a rate-limiting step in human cancer formation. As another example, certain ligand-receptor interactions are incompatible between mice and humans. Additionally, experiments have demonstrated important and significant differences in the ability to transform cells, compared with cells of murine origin. For these reasons, it remains essential to develop models of cancer that employ human cells.[3]
Targeting vectors
Isogenic cell lines are created via a process called homologous gene-targeting. Targeting vectors that utilize homologous recombination are the tools or techniques that are used to knock-in or knock-out the desired disease causing mutation or SNP (single nucleotide polymorphism) to be studied. Although disease mutations can be harvested directly from cancer patients, these cells usually contain many background mutations in addition to the specific mutation of interest, and a matched normal cell line is typically not obtained. Subsequently, targeting vectors are used to ‘knock-in’ or ‘knock out’ gene mutations enabling a switch in both directions; from a normal to cancer genotype; or vice versa; in characterized human cancer cell lines such as HCT116 or Nalm6.[4]
There are several gene targeting technologies used to engineer the desired mutation, the most prevalent of which are briefly described, including key advantages and limitations, in the summary table below.
| Technique | Gene Knock-In | Gene Knock-out |
|---|---|---|
| rAAV (recombinant adeno-associated virus vectors)[5] | Targeted insertions or modifications are created within endogenous genes; and so are subject to:
rAAV can introduce subtle point mutations, SNPs as well as small insertions with high efficiency. Moreover, many peer reviewed studies have shown that rAAV does not introduce any confounding off target genomic events. Appears to be the preferred method being adopted in academia, Biotech and Pharma on a precision versus time versus cost basis.| |
Gene knockouts are at the endogenous locus, and thus are definitive, stable and patient relevant. No confounding off-target effects are elicited at other genomic loci. It requires a 2- step process:
This process can therefore generate 3 genotypes (+/+; -/+ and -/-); enabling therefore the analysis of haplo-insufficient gene function. Current limitation is the need to sequentially target single alleles making generation of knock-out cell lines a two-step process.| |
| Plasmid based homologous recombination | Insertion is at the endogenous locus and has all the above benefits, but it is very inefficient. It also requires a promoterless drug selection strategy entailing bespoke construct generation. A large historical bank of cell-lines has been generated using this method which has been displaced by other methods since the mid 1990s. | Deletion is at endogenous locus and has all the above benefits, but it is inefficient. It also requires a promoterless drug selection strategy that entails bespoke construct generation |
| Flip-in | This is an efficient technique that allows the directed insertion of ‘ectopic’ transgenes at a single pre-defined genomic locus (integration via a FLP recombinase site). This is not a technique for modifying an endogenous locus. Transgenes will usually be under the control of an exogenous promoter, or a partially defined promoter-unit in the incorrect genomic location. Their expression will therefore not be under the same genomic and epigenetic regulation as the endogenous loci, which limits the utility of these systems for studying gene-function. They are however, good for eliciting rapid and stable exogenous gene expression. | Not applicable |
| Zinc-Finger Nucleases (ZFNs) | ZFNs have been reported to achieve high rates of genetic knock-outs within a target endogenous gene. If ZFNs are co-delivered with a transgene construct homologous to the target gene, genetic knock-in's or insertions can also be achieved.[6] One potential drawback is that any off-target double strand breaks could lead to random off-target gene insertions, deletions and wider genomic instability; confounding the resulting genotype.[7] However, no measurable increase in the rate of random plasmid integration was observed in human cells efficiently edited with ZFNs that target a composite 24 bp recognition site [6] | ZFNs are sequence-directed endonucleases which enable the rapid and highly efficient (up to 90% in a bulk cell population) disruption of both alleles of a target gene, although user- defined or patient relevant loss of-function alterations have not been reported at similar frequencies. Off target deletions or insertions elsewhere in the genome are a significant concern. The speed advantage of obtaining a biallelic KO in one step is also partially mitigated if one still needs to derive a clonal cell-line to study gene function in a homogenous cell-population. |
| Meganucleases | Meganucleases are operationally analogous to ZFN's. There are limitations inherent in their use such as the meganuclease vector design which can take up to 9 months and cost tens of thousands of dollars. This makes meganucleases more attractive in high-value applications such as gene therapy, agrobiotechnology and engineering of bioproducer lines. |
Homologous recombination in cancer cell disease models
Homologous recombination (HR) is a kind of genetic recombination in which genetic sequences are exchanged between two similar segments of DNA. HR plays a major role in eukaryotic cell division, promoting genetic diversity through the exchange between corresponding segments of DNA to create new, and potentially beneficial combinations of genes.
HR performs a second vital role in DNA repair, enabling the repair of double-strand breaks in DNA which is a common occurrence during a cell's lifecycle. It is this process which is artificially triggered by the above technologies, and bootstrapped in order to engender ‘knock-ins’ or ‘knockouts’ in specific genes5, 7.
A recent key advance was discovered using AAV-homologous recombination vectors, which increases the low natural rates of HR in differentiated human cells when combined with gene-targeting vectors-sequences.
-
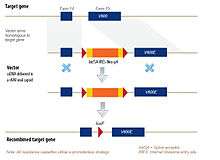
Diagram of a typical rAAV vector (source: https://www.horizondiscovery.com/gene-editing/raav)
Commercialization
Factors leading to the recent commercialization of isogenic human cancer cell disease models for the pharmaceutical industry and research laboratories are twofold.
Firstly, successful patenting of enhanced targeting vector technology has provided a basis for commercialization of the cell-models which eventuate from the application of these technologies.
Secondly, the trend of relatively low success rates in pharmaceutical RnD and the enormous costs have created a real need for new research tools that illicit how patient sub-groups will respond positively or be resistant to targeted cancer therapeutics based upon their individual genetic profile.
There are several companies working to address this need, a list of the key players and their technology offering is provided below.
- Horizon Discovery: Genesis (rAAV)
- Cellectis:Meganucleases
- Invitrogen: FLP
- Sigma-Aldrich: Zinc Fingers
See also
- AAV
- Zinc Finger Nuclease
- Plasmid
- FLP-FRT Recombination
- Homologous Recombination
- Synthetic lethality
- Recombinant AAV mediated genome engineering
- Genome engineering
News
- Masters JR (December 2000). "Human cancer cell lines: fact and fantasy". Nat. Rev. Mol. Cell Biol. 1 (3): 233–6. doi:10.1038/35043102. PMID 11252900.
- http://web.mit.edu/piyush/www/diseasemodels.pdf
- http://www.genengnews.com/gen-news-highlights/gsk-to-use-horizon-discovery-s-cell-lines-for-cancer-related-metabolomics-research/78565157/
- http://www.genomeweb.com/biotechtransferweek/horizon-discoverys-umb-cell-line-deal-latest-example-its-academic-collaboration-
- http://www.genomeweb.com/dxpgx/tgen-horizon-discovery-set-pgx-pact
- http://www.tgen.org/news/index.cfm?pageid=57&newsid=1764 TD2
- http://www.businessweekly.co.uk/life-sciences-archive/horizon-hooks-up-with-genentech.html
- http://www.horizondiscovery.com/uploads/horizon-downloads/horizon-xman-genesis-faqs.pdf /
- http://www.cellectis.com/genome-engineering/meganucleases/engineered-meganucleases/meganuclease-technologies/
- http://www.sigmaaldrich.com/life-science/zinc-finger-nuclease-technology/custom-zfn.html
- http://tools.invitrogen.com/content.cfm?pageid=3375
Sources
- Bardelli A, Parsons DW, Silliman N, et al. (May 2003). "Mutational analysis of the tyrosine kinome in colorectal cancers". Science. 300 (5621): 949. doi:10.1126/science.1082596. PMID 12738854.
- Kohli M, Rago C, Lengauer C, Kinzler KW, Vogelstein B (2004). "Facile methods for generating human somatic cell gene knockouts using recombinant adeno-associated viruses". Nucleic Acids Res. 32 (1): e3. doi:10.1093/nar/gnh009. PMC 373311
 . PMID 14704360.
. PMID 14704360. - Wang Z, Shen D, Parsons DW, et al. (May 2004). "Mutational analysis of the tyrosine phosphatome in colorectal cancers". Science. 304 (5674): 1164–6. doi:10.1126/science.1096096. PMID 15155950.
- Topaloglu O, Hurley PJ, Yildirim O, Civin CI, Bunz F (2005). "Improved methods for the generation of human gene knockout and knockin cell lines". Nucleic Acids Res. 33 (18): e158. doi:10.1093/nar/gni160. PMC 1255732
 . PMID 16214806.
. PMID 16214806. - Moroni M, Sartore-Bianchi A, Benvenuti S, Artale S, Bardelli A, Siena S (November 2005). "Somatic mutation of EGFR catalytic domain and treatment with gefitinib in colorectal cancer". Ann. Oncol. 16 (11): 1848–9. doi:10.1093/annonc/mdi356. PMID 16012179.
- Di Nicolantonio F, Bardelli A (January 2006). "Kinase mutations in cancer: chinks in the enemy's armour?". Curr Opin Oncol. 18 (1): 69–76. doi:10.1097/01.cco.0000198020.91724.48. PMID 16357567.
- Benvenuti S, Sartore-Bianchi A, Di Nicolantonio F, et al. (March 2007). "Oncogenic activation of the RAS/RAF signaling pathway impairs the response of metastatic colorectal cancers to anti-epidermal growth factor receptor antibody therapies". Cancer Res. 67 (6): 2643–8. doi:10.1158/0008-5472.CAN-06-4158. PMID 17363584.
- Arena S, Pisacane A, Mazzone M, Comoglio PM, Bardelli A (July 2007). "Genetic targeting of the kinase activity of the Met receptor in cancer cells". Proc. Natl. Acad. Sci. U.S.A. 104 (27): 11412–7. doi:10.1073/pnas.0703205104. PMC 2040912
 . PMID 17595299.
. PMID 17595299. - Konishi H, Karakas B, Abukhdeir AM, et al. (September 2007). "Knock-in of mutant K-ras in nontumorigenic human epithelial cells as a new model for studying K-ras mediated transformation". Cancer Res. 67 (18): 8460–7. doi:10.1158/0008-5472.CAN-07-0108. PMID 17875684.
- Arena S, Isella C, Martini M, de Marco A, Medico E, Bardelli A (September 2007). "Knock-in of oncogenic Kras does not transform mouse somatic cells but triggers a transcriptional response that classifies human cancers". Cancer Res. 67 (18): 8468–76. doi:10.1158/0008-5472.CAN-07-1126. PMID 17875685.
- Grim JE, Gustafson MP, Hirata RK, et al. (June 2008). "Isoform- and cell cycle-dependent substrate degradation by the Fbw7 ubiquitin ligase". J. Cell Biol. 181 (6): 913–20. doi:10.1083/jcb.200802076. PMC 2426948
 . PMID 18559665.
. PMID 18559665. - Fattah FJ, Lichter NF, Fattah KR, Oh S, Hendrickson EA (June 2008). "Ku70, an essential gene, modulates the frequency of rAAV-mediated gene targeting in human somatic cells". Proc. Natl. Acad. Sci. U.S.A. 105 (25): 8703–8. doi:10.1073/pnas.0712060105. PMC 2438404
 . PMID 18562296.
. PMID 18562296. - Di Nicolantonio F, Martini M, Molinari F, et al. (December 2008). "Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer". J. Clin. Oncol. 26 (35): 5705–12. doi:10.1200/JCO.2008.18.0786. PMID 19001320.
- Di Nicolantonio F, Arena S, Gallicchio M, et al. (December 2008). "Replacement of normal with mutant alleles in the genome of normal human cells unveils mutation-specific drug responses". Proc. Natl. Acad. Sci. U.S.A. 105 (52): 20864–9. doi:10.1073/pnas.0808757105. PMC 2634925
 . PMID 19106301.
. PMID 19106301. - Gustin JP, Karakas B, Weiss MB, et al. (February 2009). "Knockin of mutant PIK3CA activates multiple oncogenic pathways". Proc. Natl. Acad. Sci. U.S.A. 106 (8): 2835–40. doi:10.1073/pnas.0813351106. PMC 2636736
 . PMID 19196980.
. PMID 19196980. - Sartore-Bianchi A, Martini M, Molinari F, et al. (March 2009). "PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies". Cancer Res. 69 (5): 1851–7. doi:10.1158/0008-5472.CAN-08-2466. PMID 19223544.
- Sur S, Pagliarini R, Bunz F, et al. (March 2009). "A panel of isogenic human cancer cells suggests a therapeutic approach for cancers with inactivated p53". Proc. Natl. Acad. Sci. U.S.A. 106 (10): 3964–9. doi:10.1073/pnas.0813333106. PMC 2656188
 . PMID 19225112.
. PMID 19225112. - Yun J, Rago C, Cheong I, et al. (September 2009). "Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells". Science. 325 (5947): 1555–9. doi:10.1126/science.1174229. PMC 2820374
 . PMID 19661383.
. PMID 19661383. - Sartore-Bianchi A, Di Nicolantonio F, Nichelatti M, et al. (2009). Cordes N, ed. "Multi-determinants analysis of molecular alterations for predicting clinical benefit to EGFR-targeted monoclonal antibodies in colorectal cancer". PLoS ONE. 4 (10): e7287. doi:10.1371/journal.pone.0007287. PMC 2750753
 . PMID 19806185.
. PMID 19806185. - Endogenous Expression of Oncogenic PI3K Mutation Leads to Activated PI3K Signaling and an Invasive Phenotype Poster Presented at AACR/EORTC Molecular Targets and Cancer Therapeutics, Boston, USA, Nov. 2009
- Bardelli A, Siena S (March 2010). "Molecular mechanisms of resistance to cetuximab and panitumumab in colorectal cancer". J. Clin. Oncol. 28 (7): 1254–61. doi:10.1200/JCO.2009.24.6116. PMID 20100961.
- Fattah F, Lee EH, Weisensel N, Wang Y, Lichter N, Hendrickson EA (February 2010). Pearson, Christopher E., ed. "Ku regulates the non-homologous end joining pathway choice of DNA double-strand break repair in human somatic cells". PLoS Genet. 6 (2): e1000855. doi:10.1371/journal.pgen.1000855. PMC 2829059
 . PMID 20195511.
. PMID 20195511. - Buron N, Porceddu M, Brabant M, et al. (2010). Aziz SA, ed. "Use of human cancer cell lines mitochondria to explore the mechanisms of BH3 peptides and ABT-737-induced mitochondrial membrane permeabilization". PLoS ONE. 5 (3): e9924. doi:10.1371/journal.pone.0009924. PMC 2847598
 . PMID 20360986.
. PMID 20360986. - Endogenous Expression of Oncogenic PI3K Mutation Leads to accumulation of anti-apoptotic proteins in mitochondria Poster Presented at AACR 2010, Washington, D.C., USA, April. 2010
- The use of ‘X-MAN’ isogenic cell lines to define PI3-kinase inhibitor activity profiles Poster Presented at AACR 2010, Washington, D.C., USA, April. 2010
- The use of ‘X-MAN’ mutant PI3CA increases the expression of individual tubulin isoforms and promoted resistance to anti-mitotic chemotherapy drugs Poster Presented at AACR 2010, Washington, D.C., USA, April. 2010
- Di Nicolantonio F, Arena S, Tabernero J, et al. (August 2010). "Deregulation of the PI3K and KRAS signaling pathways in human cancer cells determines their response to everolimus". J. Clin. Invest. 120 (8): 2858–66. doi:10.1172/JCI37539. PMC 2912177
 . PMID 20664172.
. PMID 20664172.
References
- ↑ Torrance CJ, Agrawal V, Vogelstein B, Kinzler KW (October 2001). "Use of isogenic human cancer cells for high-throughput screening and drug discovery". Nat. Biotechnol. 19 (10): 940–5. doi:10.1038/nbt1001-940. PMID 11581659.
- ↑ "Disease models of breast cancer". Drug Discovery Today. 1: 9–16. 2004. doi:10.1016/j.ddmod.2004.05.001.
- ↑ Hirata R, Chamberlain J, Dong R, Russell DW (July 2002). "Targeted transgene insertion into human chromosomes by adeno-associated virus vectors". Nat. Biotechnol. 20 (7): 735–8. doi:10.1038/nbt0702-735. PMID 12089561.
- ↑ Masters JR (December 2000). "Human cancer cell lines: fact and fantasy". Nat. Rev. Mol. Cell Biol. 1 (3): 233–6. doi:10.1038/35043102. PMID 11252900.
- ↑ Engelhardt JF (August 2006). "AAV hits the genomic bull's-eye". Nat. Biotechnol. 24 (8): 949–50. doi:10.1038/nbt0806-949. PMID 16900138.
- 1 2 Urnov, Fyodor D.; Rebar, Edward J.; Holmes, Michael C.; Zhang, H. Steve; Gregory, Philip D. (2010). "Genome editing with engineered zinc finger nucleases". Nature Reviews Genetics. 11 (9): 636–646. doi:10.1038/nrg2842. PMID 20717154.
- ↑ Radecke S, Radecke F, Cathomen T, Schwarz K (April 2010). "Zinc-finger nuclease-induced gene repair with oligodeoxynucleotides: wanted and unwanted target locus modifications". Mol. Ther. 18 (4): 743–53. doi:10.1038/mt.2009.304. PMC 2862519
 . PMID 20068556.
. PMID 20068556.