IMES-2 RNA motif
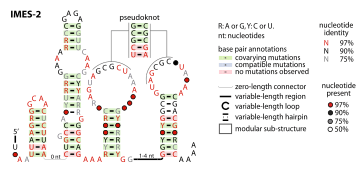
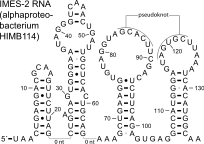
The IMES-2 RNA motif is a conserved RNA structure that was identified by a study based on metagenomics and bioinformatics,[1] and the underlying RNA sequences were identified independently by a similar earlier study.[2] These RNAs are present in environmental sequences, and when discovered were not known to be present in any cultivated species. However, an IMES-2 RNA has been detected in alphaproteobacterium HIMB114, which is classified in the SAR11 clade of marine bacteria. This finding fits with earlier predictions that species that use IMES-2 RNAs are most closely related to alphaproteobacteria.[2] IMES-2 RNAs are exceptionally abundant, as twice as many IMES-2 RNAs were found as ribosomes[1] in RNAs sampled from the Pacific Ocean.[2][3] Only two bacterial RNAs are known (6S RNA and transfer RNA) to be more highly transcribed than ribosomes.
The IMES-2 RNA secondary structure contains four stem-loop structures and one pseudoknot.
References
- 1 2 3 Weinberg Z, Perreault J, Meyer MM, Breaker RR (December 2009). "Exceptional structured noncoding RNAs revealed by bacterial metagenome analysis". Nature. 462 (7273): 656–9. doi:10.1038/nature08586. PMID 19956260.
- 1 2 3 Shi Y, Tyson GW, DeLong EF (May 2009). "Metatranscriptomics reveals unique microbial small RNAs in the ocean's water column". Nature. 459 (7244): 266–9. doi:10.1038/nature08055. PMID 19444216.
- ↑ Frias-Lopez J, Shi Y, Tyson GW, et al. (March 2008). "Microbial community gene expression in ocean surface waters". Proc. Natl. Acad. Sci. U.S.A. 105 (10): 3805–10. doi:10.1073/pnas.0708897105. PMC 2268829
 . PMID 18316740.
. PMID 18316740.